msTide
About msTide
The msTide database is a repository of negative-ion mode tandem mass spectrometry data of natural nucleotides and their synthetic derivatives. The compounds include nucleoside mono-, di- and tri- phosphates, nucleotide sugars, nucleoside phosphosulfates and their counterparts bearing modifications within the (oligo)phosphate, ribose or nucleobase. A significant group of compounds featured in the database is nucleotides modified within the phosphate with phosphorothioate, phosphoroselenoate, phosphoroamidate, boranophosphate, fluorophosphate, methylenebisphosphonate or imidodiphosphate moieties.
The database was created to accompany a research paper on the fragmentation pathways of modified nucleotides:
Strzelecka D, Chmielinski S, Bednarek S, Jemielity J, Kowalska J* Analysis of mononucleotides by tandem mass spectrometry: investigation of fragmentation pathways for phosphate- and ribose-modified nucleotide analogues, Scientific Reports, vol. 7, no. 1, Aug. 2017
*Contact corresponding author: Joanna.Kowalska AT fuw.edu.pl
Currently the database contains information on over 150 compounds. Each entry comprises the chemical structure of parent ion, precursor IUPAC name and abbreviation, measurement conditions, an MS/MS spectrum, a numerical list of registered peaks and their normalized intensities. A set of proposed chemical structures for identified ion fragments is also provided. All fragments were manually assigned and evaluated by authors of the paper cited above. In more challenging cases, additional confirmation via isotope labelling methods was obtained. MS/MS spectra were acquired at carefully adjusted collision energies and depolarization potential in order to render most comprehensive data for each compound. In all cases, soft ionization (ESI) and collision-induced dissociation (CID) were employed. Raw spectra were filtered and normalized with a cutoff set to 1% intensity.
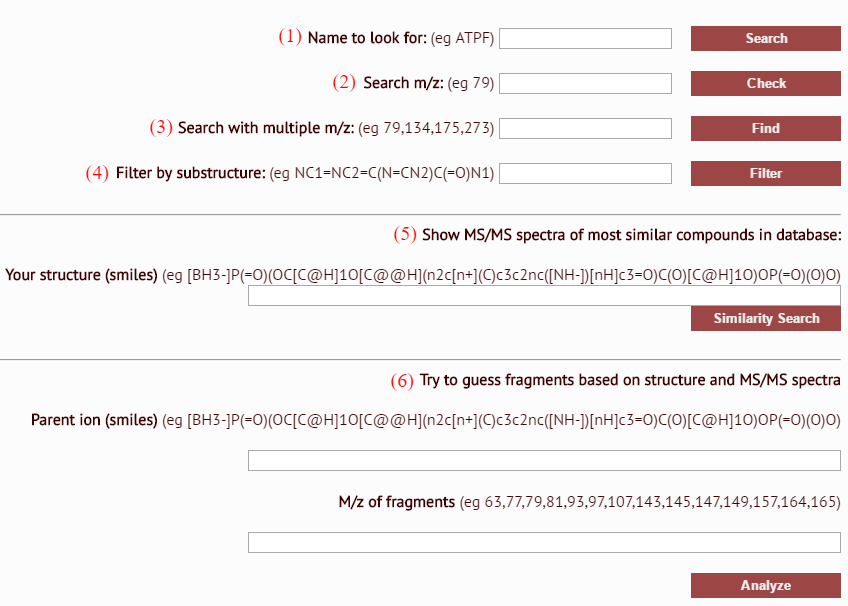
Information deposited in the database can be browsed via several search options, including monoisotopic mass search, peak (m/z) search, multiple peaks search, substructure and structural similarity search, filtering of structures by modification(s) present in the precursor.
We hope that the database would facilitate detailed analysis of nucleotide fragmentation patterns, allowing for easier identification of metabolites, synthetic products, fragmentation ions, and modifications present in nucleotides. Information stored in the database can aid development of mass spectrometry based methods for the diagnosis of nucleotide-related diseases, identification of nucleotide metabolites and quantitative analysis of selected nucleotides. It can also be applied in fast and low-material consuming identification of nucleotide-derived synthetic products. All information collected in the library can be accessed free of charge, no registration required.
How to use msTide
There are several methods for accessing information stored in the database:
- compound name search: query by IUPAC, trivial, commercial names or abbreviations (partial names can also be used)
- m/z search: generate a list of parent ions and fragment ions matching provided m/z value or molecular mass
- multiple m/z search: retrieve spectra of structures that exhibit peaks with a particular m/z values
- substructure search: retrieve a list of compounds containing provided scaffold structure (input should be in the form of canonical SMILES, which can be generated in any chemical drawing software)
- structural similarity search: retrieve a list of compounds exhibiting structural similarity to queried compound (input should be in the form of canonical SMILES, which can be generated in any chemical drawing software)
- identify MS/MS fragment ions: based on provided parent ion structure along with a list of m/z values, probable ion fragments structures will be proposed
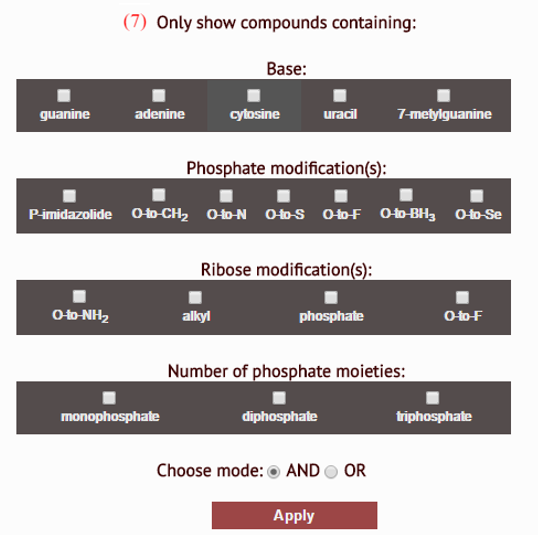
- filter compounds by modification(s) they bear: number of compounds presented for viewing can be pruned by applying one or more selection conditions, adhering to the type of base or ribose/phosphate modifications of target nucleotides
Search results are arranged in a grid, providing a way of viewing all relevant molecules at once.
Spectra comparison and Match Factor calculation
Deposited spectra can be compared for additional insight. Upon marking two compounds, user will be presented with their superimposed spectra and corresponding Match Factor. MF is a numerical value, ranging from 0 to 999, representing the degree to which two normalized MS spectra are similar. Match Factors were calculated according to the algorithms reported for the NIST library [1].
[1]S. E. Stein, “Estimating probabilities of correct identification from results of mass spectral library searches,” Journal of the American Society for Mass Spectrometry, vol. 5, no. 4, pp. 316–323, Apr. 1994.
Funding sources
The msTide database was created within the frames of a research project funded by Ministry of Science and Higher Education in Poland (IUVENTUS PLUS programme, project No IP2014 022273).